Lifespan of different species correlated with mutation rate of non-reproductive (somatic) cells. This research came out in 2022 but for some reason I only found out about it today. But it seems worth sharing as it seems a key insight into the nature of lifespan. Humans have 47 substitutions per year, while mice have 796 and live much shorter lives.
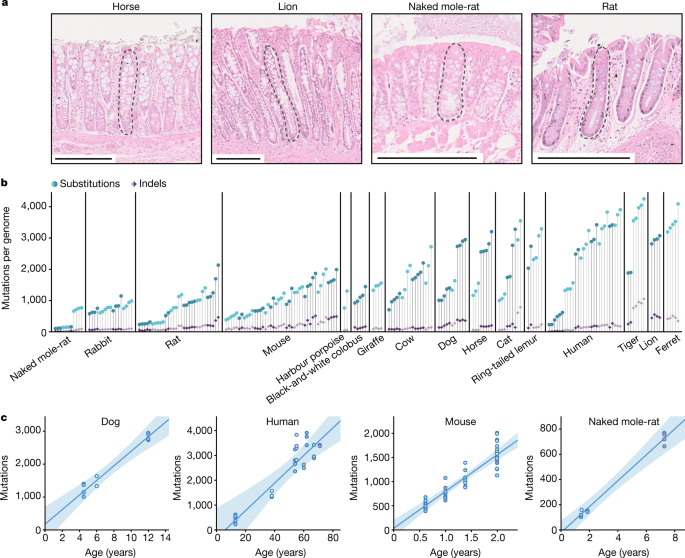
"To study somatic mutations across a diverse set of mammals, we isolated 208 individual intestinal crypts from 56 individuals across 16 species with a wide range of lifespans and body sizes: black-and-white colobus monkey, cat, cow, dog, ferret, giraffe, harbour porpoise, horse, human, lion, mouse, naked mole-rat, rabbit, rat, ring-tailed lemur, and tiger."
"We chose intestinal crypts for several reasons. First, they are histologically identifiable units that line the epithelium of the colon and small intestine and are amenable to laser microdissection. Second, human studies have confirmed that individual crypts become clonally derived from a single stem cell and show a linear accumulation of mutations with age, which enables the estimation of somatic mutation rates through genome sequencing of single crypts. Third, in most human crypts, most somatic mutations are caused by endogenous mutational processes common to other tissues, rather than by environmental mutagens."
"Across species, the mutational spectra showed clear similarities, with a dominance of cytosine-to-thymine (C>T) substitutions at CpG sites, as observed in human colon, but with considerable variation in the frequency of other substitution types."
"Across the 15 species with age information, we found that substitution rates per genome ranged from 47 substitutions per year in humans to 796 substitutions per year in mice, and indel rates from 2.5 to 158 indels per year, respectively."
"Indel", short for insertion/deletion, is a general term that may refer to any combination of insertions and deletions in DNA.
"To investigate the relationship between somatic mutation rates, lifespan and other life-history traits, we first estimated the lifespan of each species using survival curves. We used a large collection of mortality data from animals in zoos to minimize the effect of extrinsic mortality. We defined lifespan as the age at which 80% of individuals reaching adulthood have died, to reduce the effects of outliers and variable cohort sizes that affect maximum lifespan estimates. Notably, we found a tight anticorrelation between somatic mutation rates per year and lifespan across species. A log-log allometric regression yielded a strong linear anticorrelation between mutation rate per year and lifespan (fraction of inter-species variance explained = 0.85, P = 1 x 10^-6), with a slope close to and not significantly different from -1. This supports a simple model in which somatic mutation rates per year are inversely proportional to the lifespan of a species (rate is approximately equal to 1/lifespan), such that the number of somatic mutations per cell at the end of the lifespan (the end-of-lifespan burden) is similar in all species."
"To further study the relationship between somatic mutation rates and life-history variables, we used linear mixed-effects regression models. These models account for the hierarchical structure of the data (with multiple crypts per individual and multiple individuals per species), as well as the heteroscedasticity of somatic mutation rate estimates across species. Using these models, we estimated that the inverse of lifespan explained 82% of the inter-species variance in somatic substitution rates (rate = k/lifespan) (P = 2.9 x 10^-9), with the slope of this regression (k) representing the mean estimated end-of-lifespan burden across species (3,206.4 substitutions per genome per crypt, 95% confidence interval 2,683.9-3,728.9). Of note, despite uncertainty in the estimates of both somatic mutation rates and lifespans, and despite the diverse life histories of the species surveyed--including around 30-fold variation in lifespan and around 40,000-fold variation in body mass--the estimated mutation load per cell at the end of lifespan varied by only around threefold across species."
"Analogous results were obtained when repeating the analysis with estimates of the protein-coding mutation rate, which may be a better proxy for the functional effect of somatic mutations (85% of variance explained; end-of-lifespan burden: 31 coding substitutions per crypt)."
"Giraffe and naked mole-rat, for instance, have similar somatic mutation rates (99 and 93 substitutions per year, respectively), in line with their similar lifespans (80th percentiles: 24 and 25 years, respectively), despite a difference of around 23,000-fold in adult body mass. Similarly, cows, giraffes and horses weigh much more than an average human, and yet have somatic mutation rates that are several fold higher, in line with expectation from their lifespan but not their body mass."
Somatic mutation rates scale with lifespan across mammals
#discoveries #biology #genetics #dna #gwas #lifespan #longevity